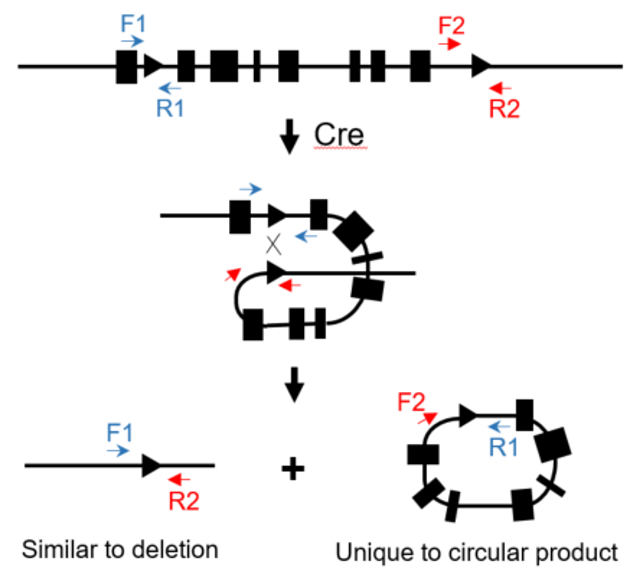
Flox: flanked by lox (sites)
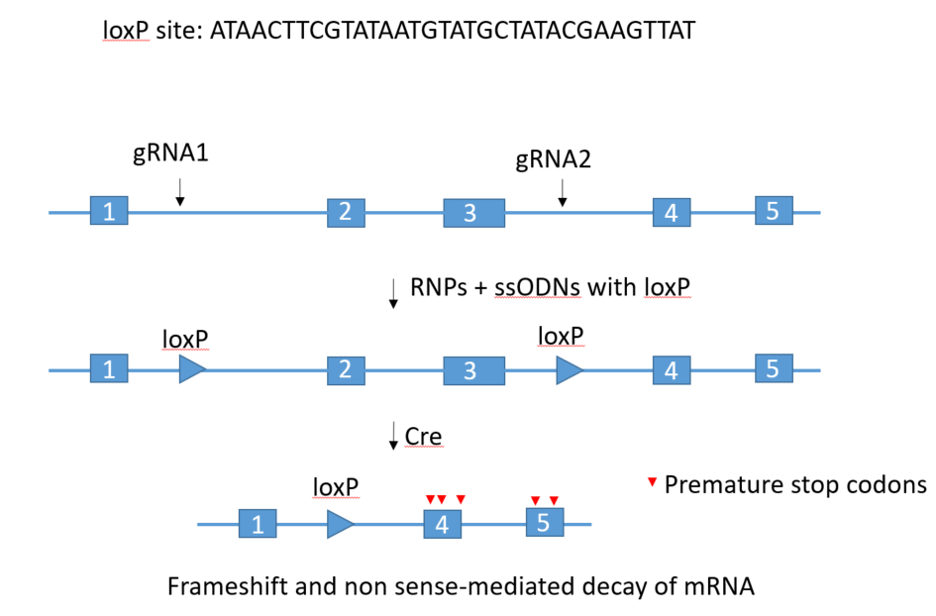
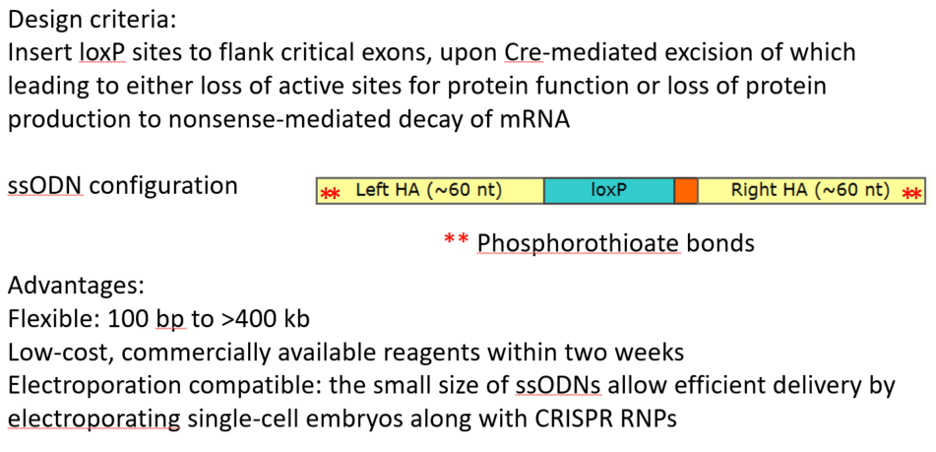
Two RNPs/two ssODNs methodology
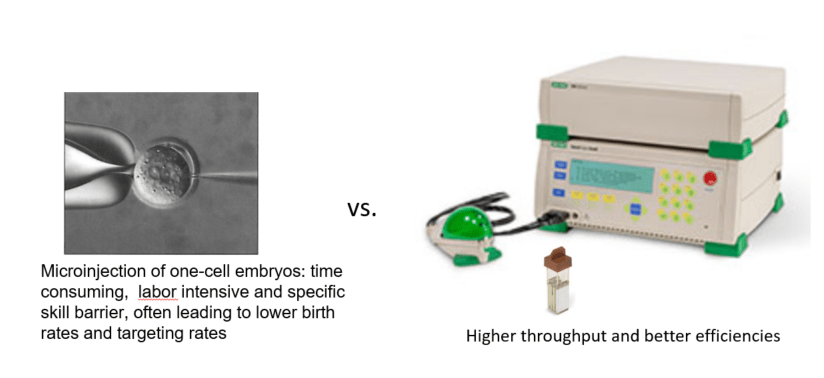
Electroporation vs. microinjection
Floxing is unlikely to be as efficient as a point mutation because of the various possible competing outcomes by exposing embryos to two gRNAs and two ssODNs
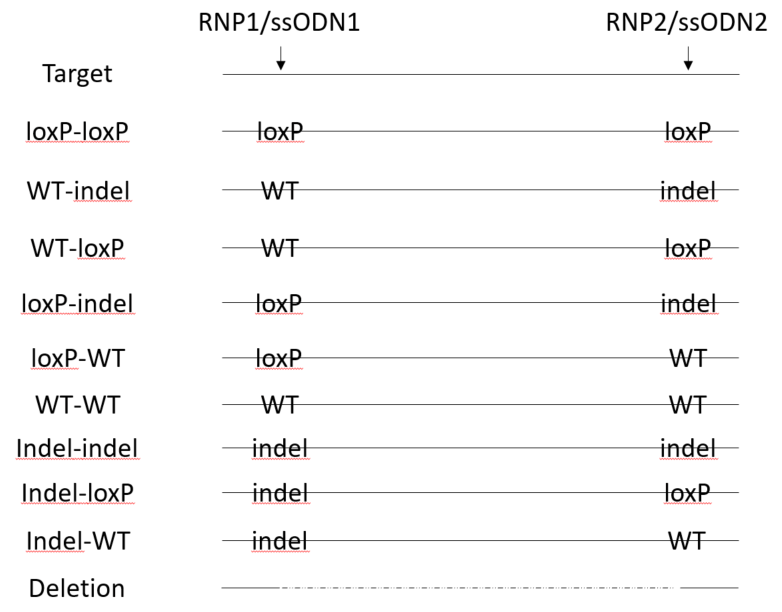
loxP insertion and indels can be detected by NGS each target site. However, deletions between the two gRNA cut sites are not amplified by small amplicons at each site. A separate PCR is needed specifically for detection of deletions

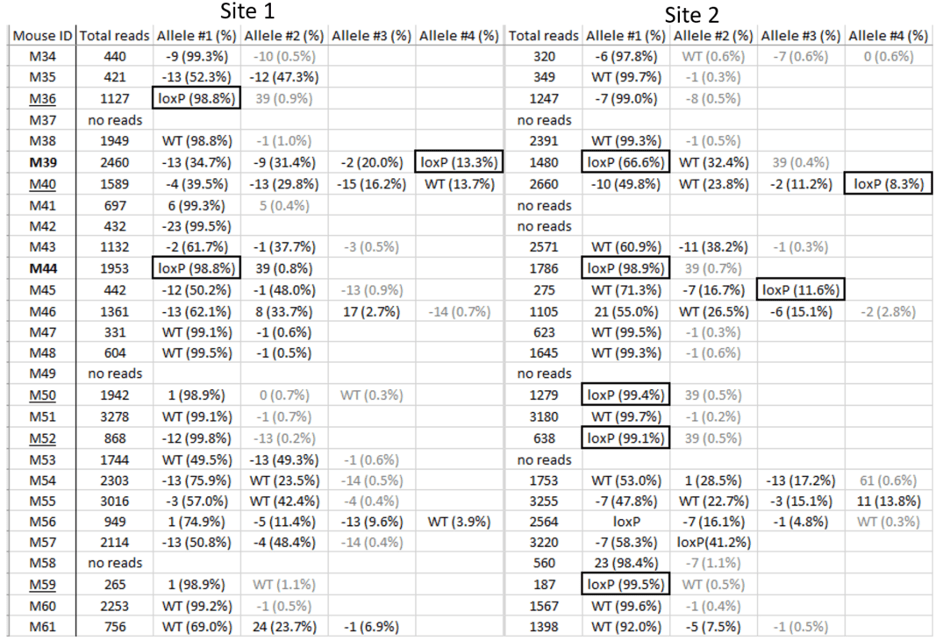
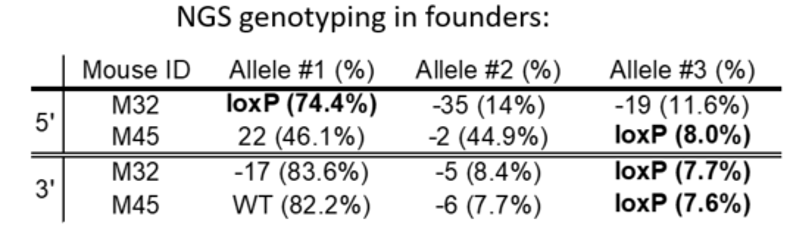
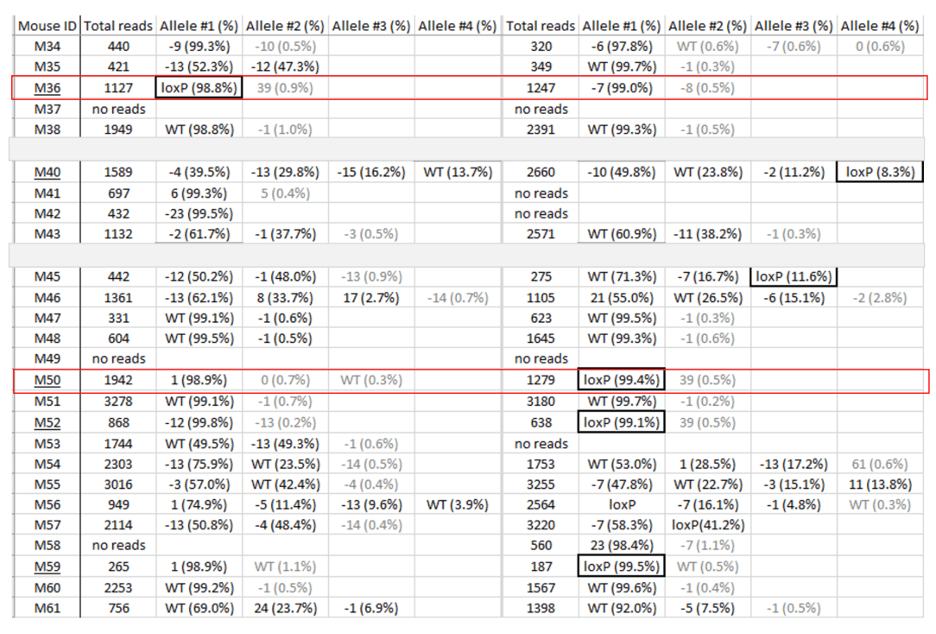
An example of genotyping results
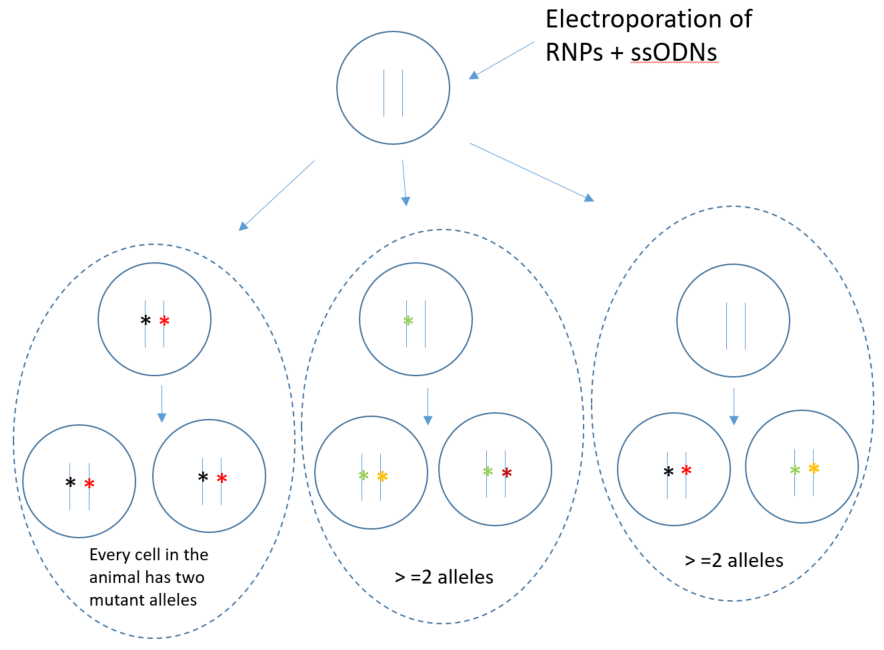
Mosaicism is common in founders born to single-cell embryo manipulation because CRIPSR RNPs last the first few cell divisions in development
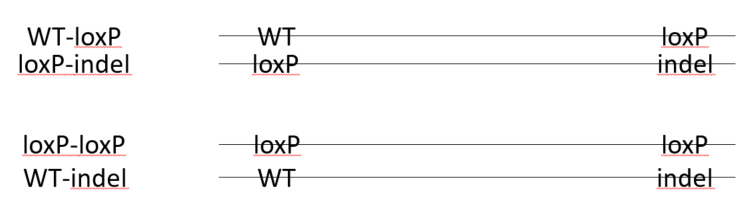
In a founder animal with loxP insertion in both target sites, it is not a guarantee that there is a floxed allele. Example of possible phasing of the loxP sites
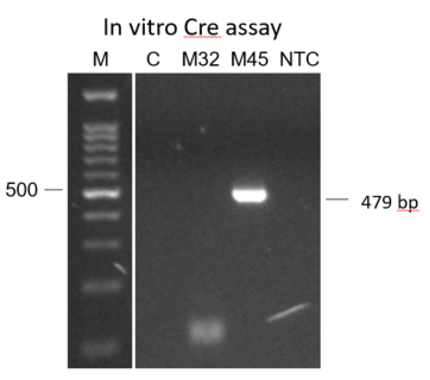
An in vitro Cre assay to determine phasing in founders

An example
What if there is no floxed founder?
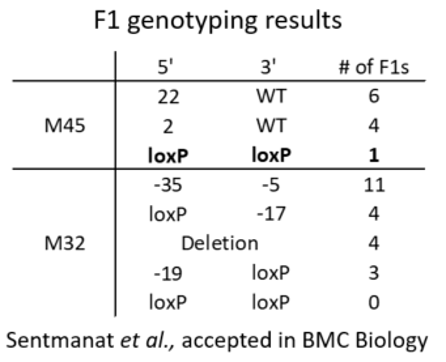
We look for founders with high percentage of loxP insertion at one site and comparably high percentage indels at the other site, see examples below in red rectangles. The rationale is the likelihood is high that those animals contain an allele with loxP/indel and a deletion allele. We can retarget the indel to insert the second loxP in embryos obtained from IVF using the sperm from these founders.
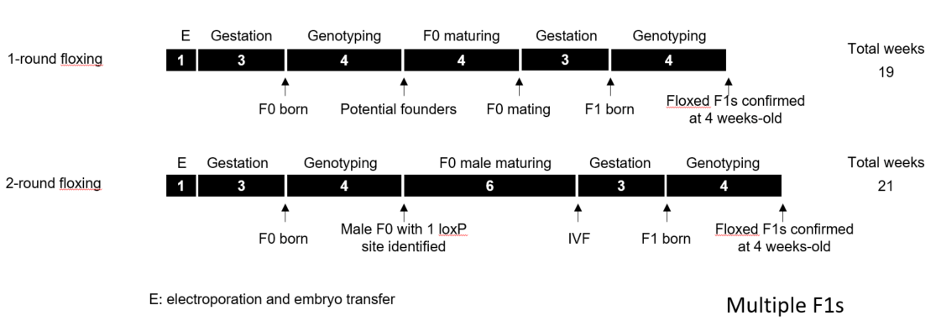
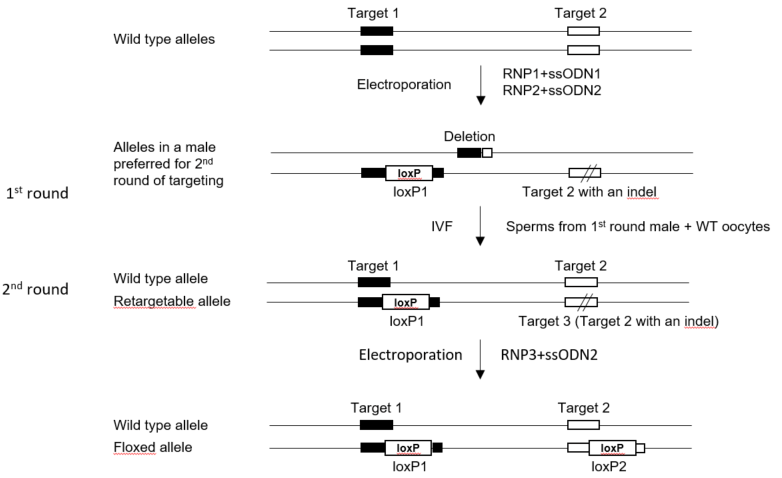
Two-round targeting strategy
Comparable timelines for 1- and 2-round strategies